21 Feb 2020
•
Technology
I recently began experimenting with spaced repetition memory systems. I was inspired to investigate this after reading Michael Nielsen’s writing on the topic (see here and here). I’m somewhat of a Michael Nielsen super-fan (see this post about a lecture he gave at Georgia Tech), so when he describes something like spaced repetition as being a superpower that radically changed his life, I have to listen closely and take heed. If you’re unfamiliar with Nielsen, I would suggest reading his Principles of Effective Research as a starting point.
Here are the tweets that set me off about spaced repetition:

(Read the entire Twitter stream here.)
I immediately downloaded Anki and began loading it up with machine learning and molecular biology questions.
In an effort to solidify my understanding of spaced repetition and get the most out of using Anki, around a year ago, I prepared a handout and gave a talk at a Junto that I frequently attend. I’m sharing that handout here in hopes that someone may find it useful.
(It was also a good excuse to try out the tufte-latex document class.)
Download the 8-page handout here:

The source is available on Github. Note that I tried to replace a lot of the examples with my own, but likely failed to attribute some ideas which were not my own.
One final note for machine learning people: Chris Albon’s famous Machine Learning Flashcards ship with an Anki-compatible set of images. Although it’s generally best to create your own cards, I have enjoyed using these in the app and highly recommend. They’re worth much more than the $12 price.
16 Feb 2020
•
Machine Learning
I started Andrew Ng’s famous Coursera Machine Learning course in 2015 after deciding to leave engineering and pursue a career in machine learning. I almost finished it, but then I actually got a job doing ML at a startup, life got crazy, and I never completed the final few weeks of the course.
I recently decided to revisit and finish it. Here are some of my thoughts:
- A large number of very well-known ML people got their start with this course. That’s pretty incredible.
- This course was exceptionally encouraging when I first started learning ML. I can’t overstate just how much Ng’s enthusiasm and encouragement increased my confidence that I could chart a new course in ML.
- The material in this course still holds up well in 2020. He doesn’t cover the latest algorithms or tools, but that’s not the point. The point of this course is to teach fundamentals and provide intuition around the foundational, classical ML algorithms.
- When I started learning ML, I found the MOOC format a nice complement to the other learning modalities I was using at the time. Some people are fine with just reading books (I read a lot of books also), but I found taking a combined approach of doing MOOCs, reading (mostly O’Reilly) books, and implementing algorithms/examples on my own very effective. Ng’s course was an important axis of my transition into ML, but by no means the whole story. I’ll write more about this another time.
- It’s really good to spend time contemplating the basics. I’ve written before (borrowing from Feynman) about how time spent on basic ideas is never wasted, and revisiting this material now was stimulating in a new way.
- Here is the course syllabus. The course provides a high level overview of linear regression, logistic regression, regularization, neural networks, support vector machines, some unsupervised learning techniques like k-means clustering and principal components analysis (PCA), basic anomaly detection approaches, collaborative filtering/recommender system basics, and a lot of more generic advice about ML system design, intuition around approaching ML problems, and techniques for improving the performance of algorithms.
- I wish the course discussed tree-based methods like decision trees, random forests, and boosting algorithms. I have always been fond of these approaches and have had good success with them. The course is time-constrained, sure, but I would have rather seen this replace support vector machines, for example.
- As someone who has been working in ML for quite a few years now, I loved hearing his anecdotal intuitions around model improvement and decision-making. I especially liked the concept of ceiling analysis, which was new to me.
- Do the programming exercises. If you have experience with linear algebra programming (Matlab, Python/numpy), the programming assignments are going to be really easy. You won’t implement any algorithms from scratch, rather you will fill in gaps in pre-written code (such as cost function, forward/backpropagation steps, etc.).
- Don’t forego the programming exercises just because they’re in Octave. Octave is extremely easy to learn and everything you need to know there can be easily googled (for loops, matrix operations, control flow).
- You’re unlikely to become an expert in any of the above just from this course, but it’s a very good orientation to the field and jumping off point.
- A lot of people wonder whether this course (or one like it) is enough to land a job in machine learning. The answer is probably “no”, and in fact for sure “no” if you have no prior technical background, don’t know linear algebra or how to program, and don’t have an existing network of software/data science people. But if you are really good at Python/math, already do technical/software/data-adjacent work, and know people, it might be enough to get your foot in the door. Although it’s not a huge time investment, completing a MOOC like this is more than nothing and can be listed on your resume/LinkedIn, unlike books you have read. If a career in ML is your goal, it’s not a bad place to start.
09 Jun 2019
•
Technology
•
Rare Disease
I have wrestled with the wisdom of having a Facebook account for years. I’ve deactivated, reactivated, and exported all my data with the intent to permanently delete (multiple times each).
I am deeply concerned about that data that companies like Facebook collect about their users. I am deeply concerned about the impact of social media on friendship as a construct. I am deeply concerned about the effect of social media on my and others’ mental health. I am deeply concerned about the impact that Facebook wields over geopolitical matters. I could link to article after article that makes coherent and compelling arguments that you should steer very clear of these technologies. I have been quite convinced by these arguments.
And so it was that in October of 2018, (I can’t remember the final straw but) I decided it was finally time to take the nuclear option and permanently delete my profile on principle.
But life was crazy and I never got around to it.
Then another thing happened on October 25, 2018: After a year of testing and investigation, we received a conclusive rare genetic disorder diagnosis for our youngest daughter M. The pediatric genetics group at Emory University was not very helpful at this point—to be fair, it’s unreasonable to expect any geneticist to know much about a disorder affecting what we would come to find out was less than 100 known individuals.
We were told, in essense: (a) Your daughter has an ultra-rare genetic disorder with fewer than 30 cases described in the medical literature, (b) we have no idea how to estimate the severity of her case and long-term prognosis, (c) come back in two years and we’ll review the literature for you to see if there’s anything new, and (d) there’s a family Facebook group you should join that might be helpful. Good luck!
The PPP2R5D Families Facebook Group
It turned out that the private PPP2R5D Families Facebook group is the central gathering point for all families worldwide who are affected by our daughter’s disorder. As soon as we saw the banner image on this Facebook page, we immediately recognized the characteristic shape of our daughter’s head in the collage of children’s photos and knew we had found our tribe.
We immediately requested to join and within hours were met with the following post in the group:
Family number 71! Our daughter M was the 71st known case in the whole world. Gina and I: 😭😭
It would be hard to put into words what this message meant to us. We had been on a 1.5-year journey at that point trying to figure out what was going on with our daughter. While our family and friends were incredibly supportive, nobody we knew got it. We didn’t know anyone who had navigated the world of #raredisease at that point. Lots of people we knew were empathetic, but almost no one we knew could actually relate.
Within 24 hours, the following things happened via this Facebook group:
- We received an outpouring of support and encouragement from families from all over the US, England, Ireland, Israel, the Netherlands, Australia, New Zealand, etc.
- We met the other family in Atlanta with a PPP2R5D variant.
- We were connected with Dr. Wendy Chung from Columbia University (PI on the 9-university research study seeking a cure), spoke with her on the phone, and were fast-tracked for an appointment in her clinic in NYC.
- We learned all about the non-profit connected to the PPP2R5D gene (Jordan’s Guardian Angels) and the coordinated research efforts underway at nine research centers all over the world.
The understanding between people in this group is unlike anything I have ever experienced. Every day someone will describe an experience or ask a question (what has worked for you for xyz symptom?) and be met with a chorus of “me too” or “here’s what worked for us” from some of the only people in the world who truly know.
Another thing that is truly incredible about this group is that not only do the families come from all over the world, but they speak different languages. The Facebook “Translate this” tool allows for reasonably seamless conversation across language barriers, even about complex medical questions.
This group of now-101 families has been a lifeline for us. As soon as we had joined this group and all the love and support started flowing in, my wife and I looked at each other and were like: Yeah, we’re not deleting Facebook.
Network effects
Facebook has also allowed for some interesting effects which we didn’t anticipate at first. After we learned about the research being conducted for PPP2R5D/Jordan’s Syndrome, we knew that we wanted to do everything we could to raise money for this nonprofit and help find a cure that would help our daughter and all other current and future children affected by this disorder.
We quickly picked up on a common tactic many families used to raise money: holding a Facebook fundraiser on the child’s birthday. M’s second birthday was approaching, and my wife Gina suggested we go for it.
This was our first foray into medical fundraising.
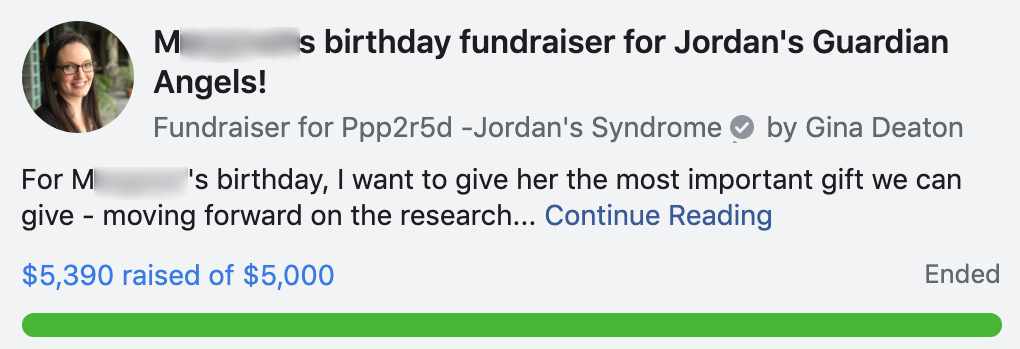
Gina set up the fundraiser and did an incredible job telling M’s story. She set it up with an initial goal of $2000, but we met that goal within 24 hours. So we increased the goal to $4000. Then we reached that goal and increased it again.
Eventually the fundraiser reached over $6000 between Facebook and direct donations to Jordan’s Guardian Angels (not captured in the image below):
Here are some things about this that surprised us:
- A huge percentage of that $6000 came from people on Facebook who (a) we don’t know very well and (b) have never even met our daughter.
- Many of our good friends shared the fundraiser with their networks which led to donations from people we don’t know at all, but who were touched by our daughter’s story.
- This is uncomfortable to discuss, but many people we know pretty well never even acknowledged the fundraiser or responded in any way. I’m not (too) upset about it, but it’s interesting to point out in the sense that if we had directly emailed people we thought would be supporters, many of the people who would have been on that list weren’t the people that ended up giving.
Facebook facilitated something here that I don’t think we could have executed otherwise.
Yes. It’s not lost on me that I have a million concerns about Facebook and then choose to participate in a group centered around the minutiae of specific genetic details pertaining to my daughter. Facebook knows the precise letters that are modified in her genome vs. the wild type human.
It is simply the price I have to pay to for access to the best information that can help her.
Alternatives to Facebook?
I’ve wracked my brain to think what an alternative gathering point would be.
What other tool meets the following criteria:
- Everyone uses it already.
- Provides built-in language translation for cross-cultural communication.
- Has public and private elements to support public dissemination of information and private collaboration of families.
- Makes it easy for advocacy and fundraising with friends who are not in the private families group.
An email list is basically the only possible alternative I can think of, and its problems seem numerous and self-evident in comparison to what Facebook enables on this front.
We could still use Facebook except with (semi-)anonymous profiles? That solves the private family group problem but not the global advocacy and fundraising aspects.
Rare disease parents don’t have time to add another tool to their arsenal or website to check. The gathering point tool has to be totally seamless or else it won’t be used.
Look, I want to hate Facebook as much as the next person but what else could accomplish everything above?
I don’t have the “luxury of opting out”, as Vicki Boykis put it recently.
What can you do?
I would appreciate it from the bottom of my heart if you would consider the following:
- Please follow PPP2R5D - Jordan’s Syndrome on Facebook to learn more about our children and the exciting research that is underway.
- Please consider supporting Jordan’s Guardian Angels, the non-profit driving the research on PPP2R5D. We have met the founders and the entire research team and can vouch for the amazing work they are doing. You can make a donation using this link that will go to directly drive the research that could help our children. 🙏
24 Apr 2019
•
Conferences
•
Data Science
Last weekend, I attended the 2019 Southern Data Science Conference (SDSC) in Atlanta. I learned a lot and met a lot of interesting people. Here are some of my takeaways.

-
I have to start by saying that Khalifeh Al Jadda and the other conference organizers did an outstanding job with the conference. This was the first single-track conference I’ve ever attended, and I appreciated the lack of anxiety involved with not having to choose between competing interests and bouncing between sessions. There were around 500 attendees, and a great mix of practitioners, researchers, and students. The whole thing was well executed.
-
The speaker lineup was impressive. Maya Gupta from Google AI, Edo Liberty, and other heads of data science/AI from Microsoft, Netflix, Pandora, Uber, LinkedIn, and many others.
-
Diversity: I would estimate over half of the keynote/panel sessions were led by women, and there was a diverse racial representation among speakers and attendees. Awesome! I also appreciated that the conference program highlighted that there would be no alcohol at the social events, which is an issue I have seen repeatedly brought up on Twitter over the years.
-
Streaming data is the future, but there are still a lot of open questions. It’s easy to think about ETL in a streaming context: recieve n new records, process them, and append them to the production cache. But there is a lot of work to be done when it comes to updating machine learning models based on a few updated observations. For now, the nightly batch job still seems in order. I am eager to work in the world of the distributed log as it seems A/B testing of feature and model changes will be much more straightforward than the way this is often done now.
-
Based on the conversations I had, most companies are still solving machine learning problems in production using one of two basic architectures: (a) Some pairing of Spark and in-memory use of scikit-learn (on either AWS or GCP) for basic regression/classification/clustering-based models, or (b) a deep learning approach using Tensorflow on GCP for NLP or computer vision. Also lots of XGBoost.
-
Serverless: Most of the people I talked to are deploying all new work on Google Cloud Platform and are migrating their legacy pipelines to GCP as quickly as they are able.
-
The poster sessions were very good. Lots of interesting ideas coming out of academia that are relevant to production machine learning work. I learned a lot from these conversations and walked away with several practical ideas, way more than I expected.
-
One of the people sitting near me had a picture of my data science friend Tim Hopper open on their computer for a few minutes (I didn’t get a close enough look to see what it was). Fun to know famous people.
-
The swag was strong. I happen to be wearing some new socks today:

All in all, it was a great several days. Will be there next year for sure.





